HOME |
| GALLERY |
| OVERVIEW |
| FAQ |
| SOURCE |
| CREDITS |
For any questions or suggestions please contact us
GoTo ...
Guidance
FastML
M1CR0B1AL1Z3R
Selecton
ConSurf
Click on the linked example titles
to view the results color-coded onto the linear or 3-dimensional structure (using
Protein Explorer) of the proteins, or click on the
static figures to see them in greater detail.
| Color coding scheme of Selecton |
| 1 2 3 4 5 6 7 |
| Positive selection Purifying selection |
Example 1: TRIM5α
Example 2: HIV-1 Protease
Example 1:
Trim5α
TRIM5 is a member of the large tripartite motif family in primate
genomes, characterized by having RING finger, B-box, and coiled-coil
domains, as well as an additional SPRY domain found in the
isoform. TRIM5 α was found to account for resistance to HIV-1 observed in
rhesus cells (Hatziioannou
et al. 2004).
TRIM5α variants from humans, rhesus monkeys and african green
monkey (AGM) display different but overlapping restriction
specificities, which all have the following common property: each TRIM5α is unable to restrict retroviruses isolated from the same species, yet is able to restrict most retroviruses from other species. This indicates that TRIM5α is an important natural barrier to cross-species retrovirus transmission.
This type of interaction between a host protein and a parasite protein
leads to genetic conflict between the two proteins. Such a conflict may
lead to rapid fixation of mutations that alter amino acids at the
protein-protein interface, i.e. positive selection. Thus, it has been
hypothesized that TRIM5α is in an antagonistic conflict with the
retroviral capsid proteins.
When a site-specific analysis was conducted,
a patch of positively selected residues was discovered in the SPRY
domain (Sawyer
et al. 2005). This patch was identified as the species-specific determinant, which is sufficient and necessary for HIV restriction in rhesus monkey cells. Substitution of this patch from the human TRIM5α with the rhesus patch, and vice-versa, conferred or abolished HIV-1 restriction, respectively . In fact, the region determining the species-specificity of the HIV-1 restriction was eventually mapped to two alternative positions in the rhesus SPRY domains. A single arginine to proline change at residue 332 of the human TRIM5α, or conversely the exchange of the six residues at position 335-340 for the eight residues of the rhesus sequence, conferred the human TRIM5α an enhanced ability to restrict HIV-1 (Sawyer
et al. 2005, Yap
et al. 2005 ).
In order to exemplify the power and ease of Selecton, the server was run
on 20 primate sequences of TRIM5α using a null model (M8a) versus
a positive selection enabling (MEC) model (p < 10-6).Results of the M8 model showed results similar to the MEC model. Figure Trim5 shows the results
of Selecton as projected onto the human primary sequence. Sites 332 and
335-340 which confer the species-specific HIV-1 restriction domain are
clearly displayed in dark yellow, indicating significant positive selection.
 |
| Fig. Trim5 |
Example 2:
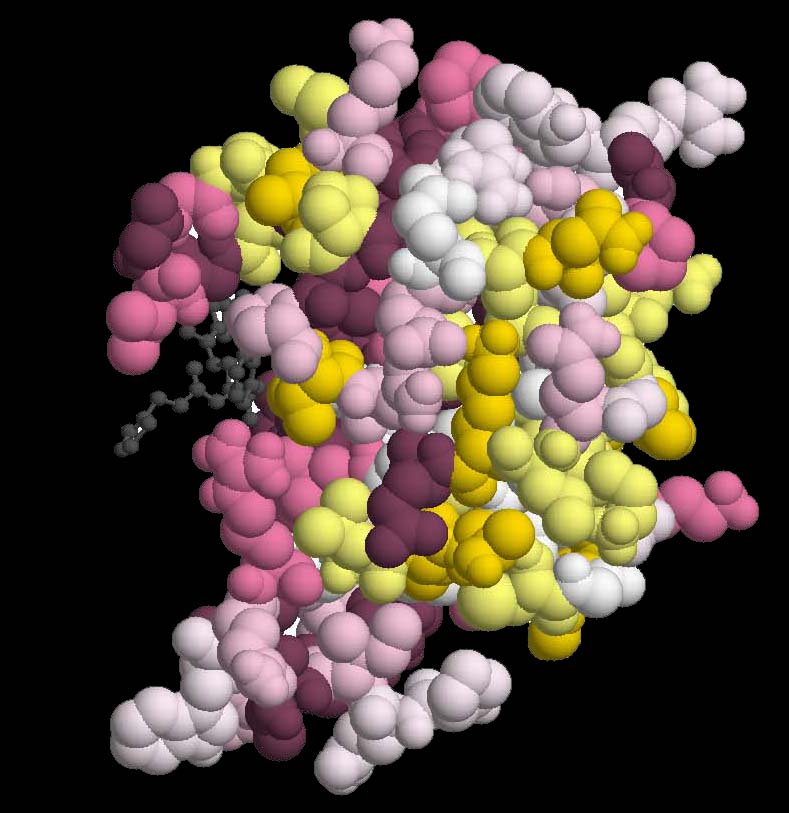
HIV-1 Protease chain A (PDB ID 1hxw chain A), Complexed With Protease Inhibitor Ritonavir.
The human immunodeficiency virus type 1 (HIV-1) protease is an essential enzyme for viral replication
and thus, is the target for design of drug inhibitors. Specific patterns of drug resistance mutations
are associated with each inhibitor available today.
Drug resistance mutations can be classified as primary when they directly confer reduced drug
susceptibility, or secondary when their influence is primarily on replication capabilities of
resistant viruses
(Lerma et al., 2001).
Seventy HIV-1 protease gene sequences from patients that were treated with a specific protease inhibitor
Ritonavir were extracted from the Stanford HIV drug resistance Database
(http://hivdb.stanford.edu/).
The sequences, together with the PDB structure of the protease dimer, complexed with Ritonavir
were given as input to the Selecton server and the results were projected onto the 3-Dimensional
structure.
Purifying selection is evident in the three known functional domains identified previously
(Loeb et al., 1989)
These domains include the active-site loop (residues 22-33), which include the Asp-Thr-Gly active-site triad characteristic of
aspartic proteases, the flap region (residues 47-52) and the hydrophobic core of the molecule (74-87) (Figure Protease_1).
Positive selected sites represent site responsible for the HIV-1 evading the treatment
conferring evolutionary advantage to the virus.
Positive selection is evident in four residues (sites 10, 54, 63, 82) where Ka/Ks is greater
than one, which have been previously reported as conferring drug resistance
(Hirsch et al., 2000)
Residue 82 which is known to belong to the primary mutation category, and is part of the
hydrophobic core, was detected by Selecton as undergoing significant positive selection.
Apart from site 82, all other previously reported sites belong to the secondary mutation category.
These sites do not fall into the three functional domains.
Top of the page